Secondary Structure
Notebook
Example Notebook: Kerasy.examples.structure.ipynb
Free energy minimization of RNA secondary structure
In cells, RNAs are likely to form energetically stable secondary structure, so the correctness of secondary structure should be evaluated based on the free energy.
Nussinov Algorithm
Nussinov Algorithm calculate the free energy by the number of base-pairs.
class kerasy.Bio.structure.Nussinov(**kwargs)
- params:
nucleic_acidWatsonCrickminspan
| variable | definition |
|---|---|
|
$$\gamma\left(i,j\right)$$
|
the maximum number of base-pairs for subsequence from \(i\) to \(j\). |
|
$$\omega\left(i,j\right)$$
|
the maximum number of base-pairs excluding subsequence from \(i\) to \(j\). |
|
$$Z(i,j)$$
|
the maximum number of base-pairs if \(x_i\) and \(y_j\) form base pair. |
$$
\begin{aligned}
\gamma(i,j) &= \max
\begin{cases}
\gamma(i+1,j)\\
\gamma(i,j-1)\\
\gamma(i+1,j-1)+\delta(i,j)\\
\max_{i\leq k\verb|<|j}\left[\gamma(i,k) + \gamma(k+1,j)\right]
\end{cases}\\
\omega(i,j) &= \max
\begin{cases}
\omega(i-1,j)\\
\omega(i,j+1)\\
\omega(i-1,j+1)+\delta(i-1,j+1)\\
\max_{1\leq k\verb|<|i}\left[\omega(k,j) + \gamma(k,i-1)\right]\\
\max_{j\verb|<| k\leq L}\left[\omega(j+1,k) + \gamma(i,k)\right]
\end{cases}\\
Z(i,j) &=
\begin{cases}
\gamma\left(i+1,j-1\right) + 1 + \omega\left(i,j\right) & (\text{if }i\text{ and }j\text{-th nucleotides can form a base-pair})\\
0 & (\text{otherwise})
\end{cases}
\end{aligned}
$$
Zuker Algorithm
class kerasy.Bio.structure.Zuker(**kwargs)
- params:
nucleic_acidWatsonCrickhairpininternalbuldgeabcstacking_colsstacking_score
The free energy of a secondary structure is approximated as the sum of the free energy of "loops".
$$E = \sum_iE_i$$
The free energy of individual loop is given by experimental data. (ex. \(\mathrm{C-G: }-3.4\mathrm{kcal/mol}\), \(\mathrm{U-A: }-0.9\mathrm{kcal/mol}\))
Five types of "loops"
| hairpin loop | stacking | bulge loop | internal loop | multi-loop |
|---|---|---|---|---|
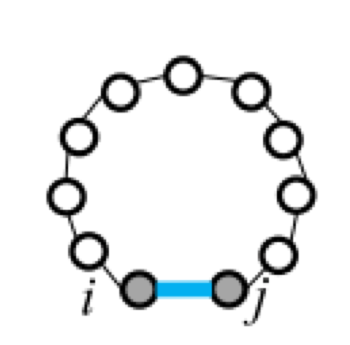 |
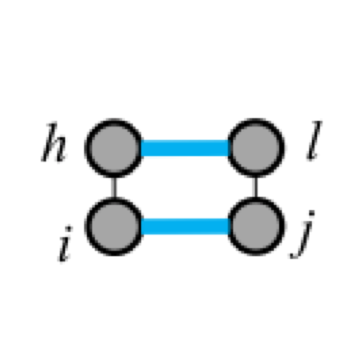 |
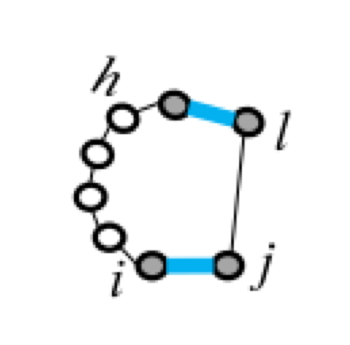 |
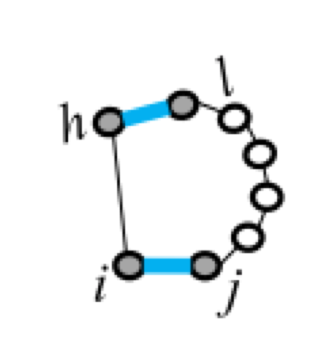 |
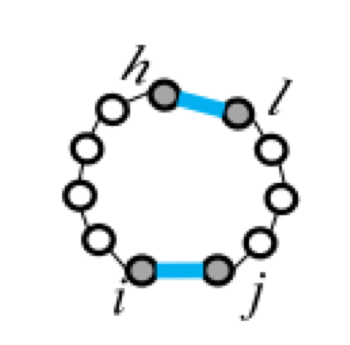 |
| F1(i,j) | F2(i,j,h,l) | Fm=a+bk+cu | ||
- a,b,c: constant
- k: the number of base-pairs in a multi-loop
- u: the number of single stranded nucleotides in a multi-loop
| variable | meaning |
|---|---|
| \(W(i,j)\) | the minimum free energy of subsequence from \(i\) to \(j\). |
| \(V(i,j)\) | the minimum free energy of subsequence from \(i\) to \(j\) when \(i\) to \(j\) forms a base-pair. |
| \(M(i,j)\) | the minimum free energy of subsequence when subsequence from \(i\) to \(j\) are in the multi-loop and contain one or more base pairs which close it. |
$$
\begin{aligned}
W(i,j) &= \min
\begin{cases}
W(i+1,j)\\W(i,j-1)\\V(i,j)\\\min_{i\leq k<j}\left\{W(i,k) + W(k+1,j)\right\}
\end{cases}\\
V(i,j) &= \min
\begin{cases}
F_1(i,j)\\\min_{i<h<l<j}F_2(i,j,h,l) + V(h,l)\\\min_{i+1\leq k<j-1}\left[M(i+1,k) + M(k+1,j)\right]+a+b
\end{cases}\\
M(i,j) &= \min
\begin{cases}
V(i,j)+b\\M(i+1,j)+c\\M(i,j-1)+c\\\min_{i\leq<k<j}\left[M(i,k) + M(k+1,j)\right]
\end{cases}\\
\end{aligned}
$$
