In [1]:
import numpy as np
import matplotlib.pyplot as plt
Section5.1 Bayesian Model Comparison¶
Overfitting¶
In [2]:
N = 15
Ms = [2,4,8,16]
seed = 0
In [3]:
# Training data.
x = np.random.RandomState(seed).uniform(low=0, high=1, size=N)
Noise = np.random.RandomState(seed).normal(loc=0, scale=0.1, size=N)
t = np.sin(2*np.pi*x) + Noise
In [4]:
fig=plt.figure(figsize=(12,8))
for i,M in enumerate(Ms):
ax=fig.add_subplot(2,2,i+1)
# Training.
X = np.array([x**m for m in range(M+1)]).T
w = np.linalg.solve(X.T.dot(X), X.T.dot(t))
# For Validation plot.
Xs = np.linspace(0,1,1000)
t_true = np.sin(2*np.pi*Xs)
t_pred = w.dot(np.array([Xs**m for m in range(M+1)]))
# RSS(Residual Sum of Squares)
residual = t - w.dot(np.array([x**m for m in range(M+1)]))
RSS = residual.T.dot(residual)
# Plot.
ax.plot(Xs,t_true,color="black", label="$t=\sin(2\pi x)$")
ax.plot(Xs,t_pred,color="red", label=f"prediction ($M={M}$)")
ax.scatter(x,t,edgecolors='blue',facecolor="white",label="train data")
ax.set_title(f"Results (RSS=${RSS:.3f}$,M=${M}$)"), ax.set_xlabel("$x$"), ax.set_ylabel("$t$")
ax.set_xlim(0,1), ax.set_ylim(-1.2,1.2)
ax.legend()
plt.tight_layout()
plt.show()
Varying Component Number¶
In [5]:
from kerasy.ML.MixedDistribution import MixedGaussian
from kerasy.utils.data_generator import generateMultivariateNormal
In [6]:
N = 200
cls = 6
seed = 123
In [7]:
data,means = generateMultivariateNormal(cls=cls, N=N, scale=8e-3, same=False, seed=seed)
x,y = data.T
mux, muy = means.T
In [8]:
# Background Color
xmin,ymin = np.min(data, axis=0); xmax,ymax = np.max(data, axis=0)
Xs,Ys = np.meshgrid(np.linspace(xmin, xmax, 100), np.linspace(ymin, ymax, 100))
XYs = np.c_[Xs.ravel(),Ys.ravel()]
In [9]:
Nfig = cls
col = 3
row = Nfig//col+1 if Nfig%col != 0 else Nfig//col
fig = plt.figure(figsize=(6*col,4*row))
Likelihoods = []
for i,K in enumerate(range(1,cls+1)):
ax = fig.add_subplot(row,col,i+1)
model = MixedGaussian(K=K, random_state=seed)
model.fit(data, span=20)
Z = model.predict(XYs)
idx = np.argmax(model.predict(data), axis=1)
Zs = np.sum(Z, axis=1).reshape(Xs.shape) # Likelihood.
likelihood = np.sum(Z)
ax.scatter(x,y,c=idx)
ax.pcolor(Xs, Ys, Zs, alpha=0.2)
ax.set_xlim(xmin, xmax)
ax.set_ylim(ymin, ymax)
ax.set_title(f"K={K}, {np.sum(Z):.1f}")
Likelihoods.append(likelihood)
plt.tight_layout()
plt.show()
In [10]:
fig = plt.figure(figsize=(12,4))
axL = fig.add_subplot(1,2,1)
axL.scatter(x,y, label="data")
axL.scatter(mux,muy, label="mean", marker="*", color="red", s=150)
axL.set_title("Training Data.")
axL.legend()
axR = fig.add_subplot(1,2,2)
axR.plot(np.arange(cls)+1,Likelihoods)
axR.scatter(np.arange(cls)+1, Likelihoods)
axR.set_xlabel("num comp")
axR.set_ylabel("likelihoods")
axR.set_title("Log likelihood vs num comp", fontsize=18)
plt.tight_layout()
plt.show()
Cross Validation¶
※ Not Bayesian
- estimate the skill of a machine learning model on unseen data.
- use a limited sample in order to estimate how the model is expected to perform in general when used to make predictions on data not used during the training of the model.
In [11]:
n_fold = 5
size = N//n_fold
Kmax = 30
In [12]:
Likelihoods = []
CrossValidationLikelihoods = []
for i,K in enumerate(range(1,Kmax+1)):
H = 0
for h in range(n_fold):
mask = np.ones(shape=N, dtype=bool)
mask[h*size:(h+1)*size] = False
model = MixedGaussian(K=K, random_state=seed)
model.fit(data[mask], span=20)
Z = model.predict(data[np.logical_not(mask)])
H += np.sum(Z)
model = MixedGaussian(K=K, random_state=seed)
model.fit(data, span=20)
Z = model.predict(data)
Likelihoods.append(np.sum(Z))
CrossValidationLikelihoods.append(H)
In [13]:
fig = plt.figure(figsize=(12,4))
axL = fig.add_subplot(1,2,1)
axL.plot(np.arange(Kmax)+1, CrossValidationLikelihoods)
axL.scatter(np.arange(Kmax)+1, CrossValidationLikelihoods)
axL.set_xlabel("num comp"), axL.set_ylabel("Cross Validation likelihoods"), axL.set_title("Cross Validation Log likelihood vs num comp", fontsize=18)
axR = fig.add_subplot(1,2,2)
axR.plot(np.arange(Kmax)+1, Likelihoods)
axR.scatter(np.arange(Kmax)+1, Likelihoods)
axR.set_xlabel("num comp"), axR.set_ylabel("likelihoods"), axR.set_title("Log likelihood vs num comp", fontsize=18)
plt.tight_layout()
plt.show()
Cross Entropy of Prediction¶
※ Not Bayesian
empirical distribution¶
- $D = \left\{\mathbf{x}^{(h)}|h=1,\ldots,n\right\}$
- $x\sim\mathrm{Emp}(D)$
- $f_X(\mathbf{X}) = \frac{1}{n}\sum_{h=1}^n\delta\left(\mathbf{x}-\mathbf{x}^{(h)}\right)$
- $\mathbb{E}\left(g(\mathbf{x})\right) = \frac{1}{n}\sum_{h=1}^n\int d\mathbf{x}g(\mathbf{x})\delta\left(\mathbf{x}-\mathbf{x}^{(h)}\right) = \frac{1}{n}\sum_{h=1}^ng\left(\mathbf{x}^{(h)}\right)$
In [14]:
seed = 0
N = 1000
In [15]:
data = np.random.RandomState(seed).normal(size=N)
In [16]:
Emp = dict(zip(data,np.zeros(N)))
for x in data: Emp[x] += 1/N
In [17]:
for X,Y in sorted(Emp.items()):
plt.plot((X,X),(0,Y), color="black", alpha=0.1)
plt.title("Empirical distribution (Normal)")
plt.xlabel("x")
plt.show()
We can assume Empirical distribution as a true distribution.
Cross Entropy¶
$$
\begin{aligned}
H(D,\boldsymbol{\theta})
&= \text{Cross Entropy between }f_{\text{Emp}(D)} \text{ and }f\left(\mathbf{x}|\boldsymbol{\theta}\right)\\
&= \mathbb{E}_{\text{Emp}(D)}\left(-\log f\left(\mathbf{X}|\boldsymbol{\theta}\right)\right)\\
&= -\int f_{\text{Emp}(D)}(\mathbf{x})\log f\left(\mathbf{x}|\boldsymbol{\theta}\right)d\mathbf{x}\\
&= -\frac{1}{n}\sum_{h=1}^n\log f\left(\mathbf{x}^{(h)}|\boldsymbol{\theta}\right)\\
&= -\bar{l}^{(n)}\left(\mathbf{x}|\boldsymbol{\theta}\right)
\end{aligned}
$$
We can also say that:
$$ \begin{aligned} H(D,\boldsymbol{\theta}) &= -\int f_{\text{Emp}}(\mathbf{x})\log f\left(\mathbf{x}|\boldsymbol{\theta}\right)d\mathbf{x}\\ &\Rightarrow \int f_{\text{Emp}}(\mathbf{x})\log f_{\text{Emp}}(\mathbf{x})d\mathbf{x} -\int f_{\text{Emp}}(\mathbf{x})\log f\left(\mathbf{x}|\boldsymbol{\theta}\right)d\mathbf{x}\\ &= -\int f_{\text{Emp}}(\mathbf{x})\log\left\{\frac{f_{\text{Emp}}(\mathbf{x})}{f\left(\mathbf{x}|\boldsymbol{\theta}\right)}\right\}d\mathbf{x}\\ &= \mathrm{KL}\left(f_{\text{Emp}}\|f\left(\mathbf{x}|\boldsymbol{\theta}\right)\right)\\ &= \text{Kullback-Leibler divergence between}f_{\text{Emp}(D)} \text{ and }f\left(\mathbf{x}|\boldsymbol{\theta}\right) \end{aligned} $$Evidence Approximation¶
※ Yes Bayesian
- In a fully Bayesian treatment of the linear basis function model, we would introduce prior distributions over the hyperparameters $\alpha$, and $\beta$. $$ \begin{aligned} p(t|\mathbf{t},\mathbf{X},\mathbf{x},\alpha,\beta) &= \int p(t|\mathbf{w},\mathbf{x},\beta)p(\mathbf{w}|\mathbf{t},\mathbf{X},\alpha,\beta)d\mathbf{w}& (3.57)\\ &\Rightarrow \int p(t|\mathbf{w},\mathbf{x},\beta)p(\mathbf{w}|\mathbf{t},\mathbf{X},\alpha,\beta)p(\alpha,\beta|\mathbf{t},\mathbf{X})d\mathbf{w}d\alpha d\beta& (3.74) \end{aligned} $$
- Marginalize with respect to these hyperparameters as well as with respect to the parameters $\mathbf{w}$ to make predictions.
- Complete marginalization over all of these variables is analytically intractable.
- We will maximize $(3.74)$ in line with the framework of empirical Bayes (or type 2 maximum likelihood, generalized maximum likelihood, evidence approximation)
- Obtain the marginal likelihoo function by first integrating over the parameters $\mathbf{w}$ $$p(\mathbf{t}|\alpha,\beta) = \int p(\mathbf{t}|\mathbf{w},\mathbf{X},\beta)p(\mathbf{w}|\alpha)d\mathbf{w}\qquad (3.77)$$
- Maximize $p(\mathbf{t}|\alpha,\beta)$ with respect to $\alpha$ and $\beta$.
In [18]:
from kerasy.utils.preprocessing import PolynomialBaseTransformer
from kerasy.utils.data_generator import generateSin, generateGausian
from kerasy.ML.linear import EvidenceApproxBayesianRegression
In [19]:
N = 20
Ms = np.arange(10)
xmin=0; xmax=1
seed = 0
In [20]:
x_train,y_train = generateSin(size=N, xmin=xmin,xmax=xmax,seed=seed)
_, y_noise = generateGausian(size=N, x=x_train, loc=0, scale=0.15, seed=seed)
y_train += 1+y_noise
In [21]:
alpha = 100
beta = 100
In [22]:
evidences = []
history = []
for M in Ms:
phi = PolynomialBaseTransformer(M=M)
x_train_features = phi.transform(x_train)
model = EvidenceApproxBayesianRegression(alpha=alpha, beta=beta, iter_max=10000)
model.fit(x_train_features,y_train)
evidences.append(model.evidence(x_train_features, y_train))
history.append(model)
best_M = np.argmax(evidences[1:])+1
best_model = history[best_M]
In [23]:
x_test = np.linspace(xmin,xmax,1000)
phi = PolynomialBaseTransformer(M=best_M)
x_test_features = phi.transform(x_test)
y_test = 1+np.sin(2*np.pi*x_test)
y_pred,y_std = best_model.predict(x_test_features)
In [24]:
fig = plt.figure(figsize=(12,4))
axEVI = fig.add_subplot(1,2,1)
axEVI.plot(Ms, evidences)
axEVI.scatter(Ms, evidences)
axEVI.set_title("The relationship between 'model evidence' and $M$"), axEVI.set_xlabel("$M$"),axEVI.set_ylabel("$\log({model\ evidence})$", fontsize=16)
axBest = fig.add_subplot(1,2,2)
axBest.plot(x_test,y_test,color="black", label="$\sin(2\pi x)$")
axBest.plot(x_test,y_pred,color="red", label="predict($\mu$)")
axBest.fill_between(x_test, y_pred-y_std, y_pred+y_std, color="pink", alpha=0.5)
axBest.scatter(x_train,y_train,s=50,edgecolors='black',facecolor="white",label="train data")
axBest.set_ylim(-0.5,2.5), axBest.legend(), axBest.set_title(f"Bayesian Linear Regression (M={best_M})")
plt.tight_layout()
plt.show()
In [25]:
print("[Best Parameter]")
print(f"M = {best_M}")
print(f"alpha = {best_model.alpha}")
print(f"beta = {best_model.beta}")
Section5.2 Markov Process¶
- Biological Diversity
- Single Origin of Life
- "Nothing in Biology Makes Sense Except in the Light of Evolution" C.T. Dobzhansky
- Stochastic Transcription (Stochasticity vs Regulation)
- Stochasticity in Development
Stochastic Process¶
Mathematical description of stochastic behaviors.
- data: $\left\{\mathbf{X}_t|t=1,\ldots,n\right\}$
- Probability: $\mathbb{P}\left(\mathbf{X}_{t_m} = \mathbf{x}_{t_m},\ldots,\mathbf{X}_{t_2} = \mathbf{x}_{t_2},\mathbf{X}_{t_1} = \mathbf{x}_{t_1}\right)$
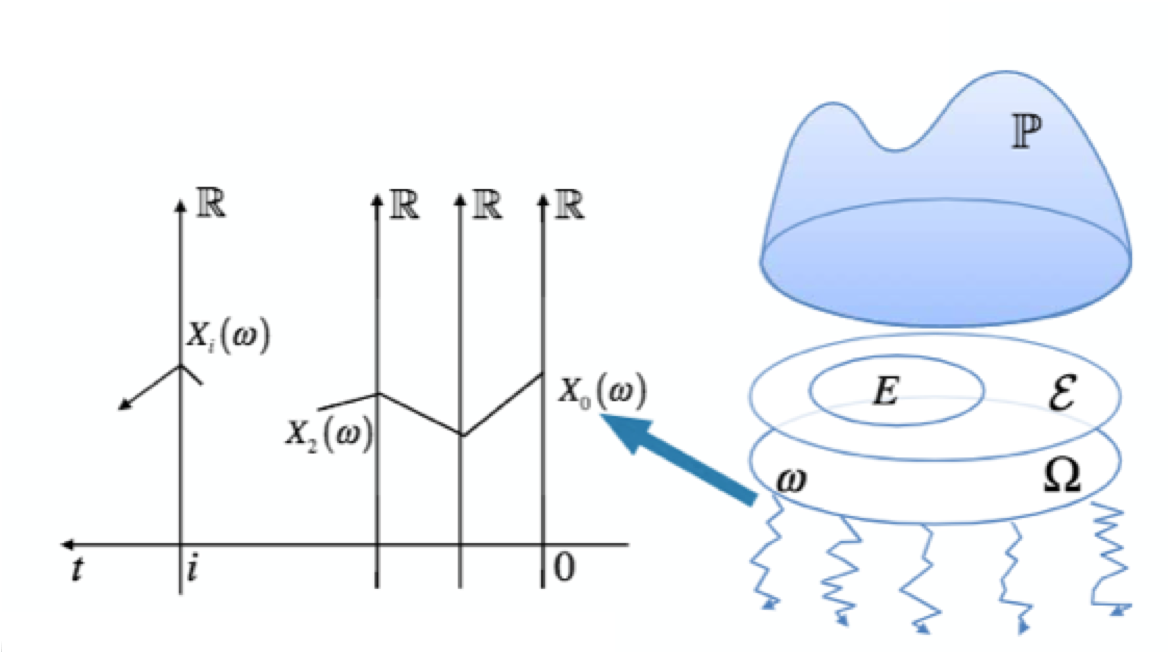
- $\omega$ is one of the random paths.
- $\mathbf{X}_t$ is a map indicats which state random paths $\omega$ is in at time $t$
- Events $E$ is a constraint on the value of $\mathbf{X}_t$ at some times.
- $P(E)$ is the probability if the random paths satisfy the constraint $E$.
markov Process¶
$$\mathbb{P}\left(\mathbf{X}_t = \mathbf{x}_t|\mathbf{X}_{t-1} = \mathbf{x}_{t-1},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right) = \mathbb{P}\left(\mathbf{X}_t=\mathbf{x}_t|\mathbf{X}_{t-1}=\mathbf{x_{t-1}}\right)$$- Decomposition of Joint Distribution $$ \begin{aligned} &\mathbb{P}\left(\mathbf{X}_t = \mathbf{x}_t,\mathbf{X}_{t-1} = \mathbf{x}_{t-1},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\\ &=\mathbb{P}\left(\mathbf{X}_t = \mathbf{x}_t | \mathbf{X}_{t-1} = \mathbf{x}_{t-1},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\times\mathbb{P}\left(\mathbf{X}_{t-1} = \mathbf{x}_{t-1},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\\ &=\mathbb{P}\left(\mathbf{X}_t = \mathbf{x}_t | \mathbf{X}_{t-1} = \mathbf{x}_{t-1},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\times \mathbb{P}\left(\mathbf{X}_{t-1} = \mathbf{x}_{t-1} | \mathbf{X}_{t-2} = \mathbf{x}_{t-2},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\times \mathbb{P}\left(\mathbf{X}_{t-2} = \mathbf{x}_{t-2},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\\ &= \mathbb{P}\left(\mathbf{X}_t = \mathbf{x}_t | \mathbf{X}_{t-1} = \mathbf{x}_{t-1},\ldots,\mathbf{X}_0 = \mathbf{x}_0\right)\times \cdots \times\mathbb{P}\left(\mathbf{X}_2=\mathbf{x}_2|\mathbf{X}_1=\mathbf{x}_1,\mathbf{X}_0=\mathbf{x}_0\right)\times \mathbb{P}\left(\mathbf{X}_1=\mathbf{x}_1|\mathbf{X}_0=\mathbf{x}_0\right)\times\mathbb{P}\left(\mathbf{X}_0=\mathbf{x}_0\right)\\ &= \mathbb{P}\left(\mathbf{X}_t = \mathbf{x}_t | \mathbf{X}_{t-1} = \mathbf{x}_{t-1}\right) \times \mathbb{P}\left(\mathbf{X}_{t-1} = \mathbf{x}_{t-1} | \mathbf{X}_{t-2} = \mathbf{x}_{t-2}\right)\times \cdots\times\mathbb{P}\left(\mathbf{X}_1=\mathbf{x}_1|\mathbf{X}_0=\mathbf{x}_0\right)\times\mathbb{P}\left(\mathbf{X}_0=\mathbf{x}_0\right)\quad\left(\because \text{Markov Process}\right) \end{aligned} $$
- Transition Matrix (Assume Time Independence): $$P = \left\{P_{ij}\right\} = \left\{\mathbb{P}\left(\mathbf{X}_t=i|\mathbf{X}_{t-1}=j\right)\right\},\quad P_{ij}\geq0,\sum_i P_{ij}=1$$
Computing transition probability¶
- Birth and Death process $$ P= \left(\begin{matrix}1-\lambda_0&\mu_1&0&0\\\lambda_0&1-\lambda_1-\mu_1&\mu_2&0\\0&\lambda_1&1-\lambda_2-\mu_2&\mu_3\\0&0&\lambda_2&1-\mu_3\end{matrix}\right) $$
- $P = U\Lambda U^{-1}$: Eigen decomposition
- $P^n = U\Lambda^n U^{-1}$
In [26]:
def makeRandomP(N, seed=123):
lambdas = np.r_[np.random.RandomState(seed).uniform(low=0,high=1/N, size=N-1), np.zeros(shape=1)]
mus = np.r_[np.zeros(shape=1), np.random.RandomState(seed+1).uniform(low=0,high=1/N, size=N-1)]
P = np.zeros(shape=(N,N))
for i in range(N):
P[i,-1+i] = mus[i];
P[i,i] = 1-mus[i]-lambdas[i];
P[i,1+i if 1+i<N else 1+i-N] = lambdas[i]
return P
In [27]:
def LongTimeBehaviors(P,even=True):
inf=50 if even else 51
eigenvals,eigenvecs = np.linalg.eig(P)
Lambda = np.diag(eigenvals)
eigenvecs_inv = np.linalg.inv(eigenvecs)
Pn = np.dot(eigenvecs, np.dot(Lambda**inf, eigenvecs_inv))
Pn = np.where(Pn<1e-5, 0, Pn)
return Pn
example¶
Periodic¶
In [28]:
P = np.array([
[0, 1, 0],
[1/2, 0, 1/2],
[0, 1, 0]
])
In [29]:
print(f"P^2n: \n{LongTimeBehaviors(P, even=True)}\n")
print(f"P^2n+1: \n{LongTimeBehaviors(P, even=False)}")
Reducible¶
In [30]:
P = np.array([
[1/3, 3/4, 0, 0],
[2/3, 1/4, 0, 0],
[0, 0, 1/3, 3/4],
[0, 0, 2/3, 1/4]
])
In [31]:
LongTimeBehaviors(P)
Out[31]:
Absorbing¶
In [32]:
P = np.array([
[1, 1/2, 0, 0],
[0, 0, 1/2, 0],
[0, 1/2, 0, 0],
[0, 0, 1/2, 1]
])
In [33]:
LongTimeBehaviors(P)
Out[33]:
Irreducible, Aperiodic¶
In [34]:
P = np.array([
[2/3, 1/12, 0],
[1/3, 5/8, 1/8],
[0, 7/24, 7/8],
])
In [35]:
LongTimeBehaviors(P)
Out[35]:
In [ ]: